Navigating the Complexities of the PT Compact Map: A Comprehensive Guide
Related Articles: Navigating the Complexities of the PT Compact Map: A Comprehensive Guide
Introduction
In this auspicious occasion, we are delighted to delve into the intriguing topic related to Navigating the Complexities of the PT Compact Map: A Comprehensive Guide. Let’s weave interesting information and offer fresh perspectives to the readers.
Table of Content
- 1 Related Articles: Navigating the Complexities of the PT Compact Map: A Comprehensive Guide
- 2 Introduction
- 3 Navigating the Complexities of the PT Compact Map: A Comprehensive Guide
- 3.1 Understanding the PT Compact Map: A Deep Dive
- 3.2 The Significance of the PT Compact Map: Unveiling Protein Evolution and Design
- 3.3 Applications of the PT Compact Map: A Glimpse into its Impact
- 3.4 Frequently Asked Questions (FAQs)
- 3.5 Tips for Using the PT Compact Map
- 3.6 Conclusion
- 4 Closure
Navigating the Complexities of the PT Compact Map: A Comprehensive Guide
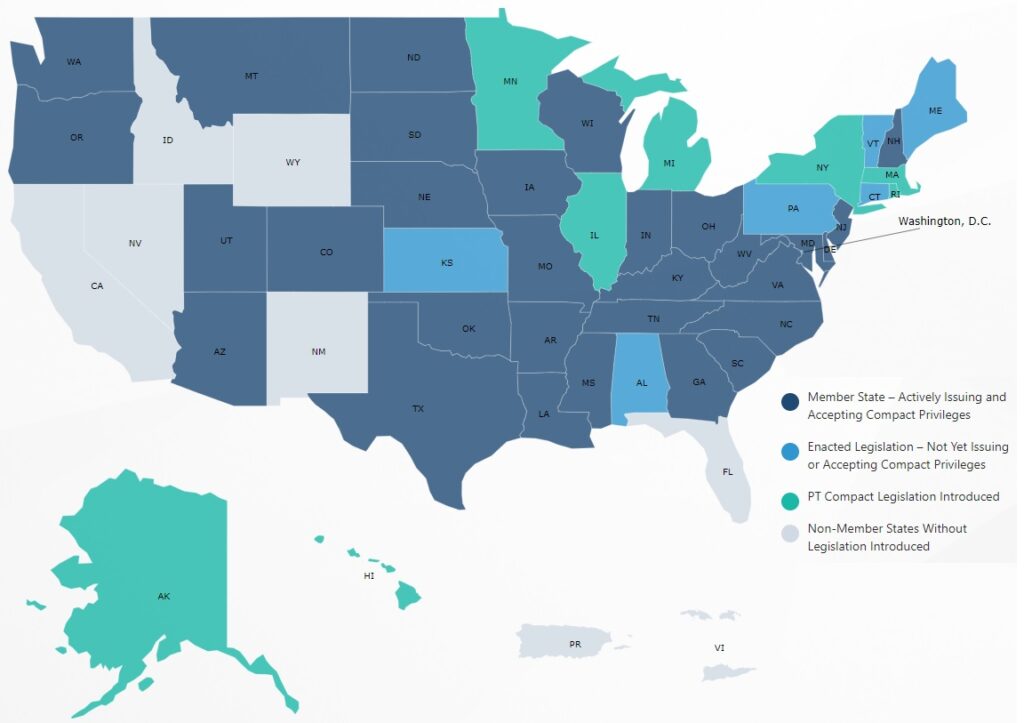
The PT Compact Map, a tool employed in the realm of protein engineering, serves as a visual representation of the evolutionary relationships between different protein families. This map, meticulously crafted through bioinformatics analysis, offers a unique lens for understanding protein structure, function, and evolution. By providing a comprehensive overview of protein families and their interconnections, the PT Compact Map empowers researchers to navigate the vast landscape of protein diversity, facilitating the design of novel proteins with tailored properties.
Understanding the PT Compact Map: A Deep Dive
The PT Compact Map is a graphical representation of the protein space, built upon the concept of protein families. Protein families are groups of proteins sharing significant structural and functional similarities, often stemming from a common evolutionary ancestor. Each protein family on the map is represented by a node, while the connections between these nodes depict the evolutionary relationships between families.
Key Components of the PT Compact Map:
- Nodes: Each node represents a distinct protein family, characterized by specific structural and functional features.
- Edges: Edges connecting nodes signify evolutionary relationships between protein families. The thickness of the edge often reflects the degree of similarity between families.
- Color Coding: Different colors are used to represent various protein families, aiding in visual identification and categorization.
The Significance of the PT Compact Map: Unveiling Protein Evolution and Design
The PT Compact Map plays a pivotal role in several areas of protein research, including:
1. Understanding Protein Evolution:
- The map facilitates the tracing of protein evolution, allowing researchers to reconstruct the evolutionary history of protein families.
- It reveals the intricate web of relationships between protein families, shedding light on how proteins have diversified over time.
- This knowledge empowers researchers to understand the mechanisms driving protein evolution, including gene duplication, gene fusion, and horizontal gene transfer.
2. Identifying Novel Protein Families:
- The map provides a framework for identifying novel protein families, which may possess unique structural and functional properties.
- By analyzing the proximity of newly discovered proteins to existing families on the map, researchers can gain insights into their potential roles and functions.
- This discovery process can lead to the identification of proteins with potential applications in various fields, including medicine, biotechnology, and materials science.
3. Designing New Proteins:
- The map serves as a guide for designing new proteins with desired properties.
- By understanding the evolutionary relationships between families, researchers can identify potential candidates for protein engineering projects.
- The map can also be used to predict the functional consequences of mutations, aiding in the design of proteins with enhanced stability, activity, or specificity.
4. Exploring Protein Function:
- The map provides a framework for exploring the functional diversity of protein families.
- By analyzing the distribution of functional annotations within families, researchers can gain insights into the evolution of protein function.
- This knowledge can be leveraged to predict the functions of newly discovered proteins or to design proteins with specific functional capabilities.
Applications of the PT Compact Map: A Glimpse into its Impact
The PT Compact Map has found applications in a wide range of research areas, including:
- Drug Discovery: The map aids in the identification of potential drug targets by highlighting proteins with unique structural and functional features.
- Biotechnology: The map facilitates the design of enzymes with enhanced activity or stability for industrial applications.
- Materials Science: The map can be used to design proteins with specific properties for the development of novel materials.
- Evolutionary Biology: The map provides insights into the evolutionary history of protein families, contributing to our understanding of the evolution of life.
Frequently Asked Questions (FAQs)
Q: What is the difference between a protein family and a protein domain?
A: A protein family is a group of proteins sharing significant structural and functional similarities, often stemming from a common evolutionary ancestor. A protein domain, on the other hand, is a distinct structural and functional unit within a protein, which can be found in multiple proteins, even those belonging to different families.
Q: How is the PT Compact Map constructed?
A: The PT Compact Map is constructed using bioinformatics techniques, analyzing large datasets of protein sequences and structures. The map is built by comparing protein sequences and identifying regions of similarity, which are then used to infer evolutionary relationships between protein families.
Q: What are the limitations of the PT Compact Map?
A: The PT Compact Map is a valuable tool but has limitations. It is based on sequence similarity, which may not always reflect true evolutionary relationships. Additionally, the map does not account for all protein families, particularly those with limited sequence information.
Tips for Using the PT Compact Map
- Familiarize yourself with the map’s structure and terminology.
- Use the map to identify potential protein families for research projects.
- Explore the map’s connections to understand evolutionary relationships between families.
- Combine the map with other data sources, such as structural information and functional annotations.
Conclusion
The PT Compact Map serves as a powerful tool for understanding the complex world of proteins. By visualizing the evolutionary relationships between protein families, it provides a framework for exploring protein structure, function, and evolution. Its applications extend across diverse fields, from drug discovery to materials science, highlighting its significance in advancing our understanding of protein diversity and its potential for innovation. As our knowledge of protein families continues to grow, the PT Compact Map will undoubtedly remain a valuable resource for researchers seeking to unravel the secrets of the protein world.





Closure
Thus, we hope this article has provided valuable insights into Navigating the Complexities of the PT Compact Map: A Comprehensive Guide. We hope you find this article informative and beneficial. See you in our next article!